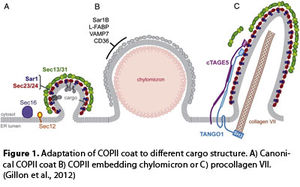
The initial step of the vesicular trafficking is vesicle budding from the Endoplasmic Reticulum (ER) that is fundamental for the structural and functional maintenance of post ER compartments and for the secretion of extracellular factors like hormones or extracellular matrix components. Vesicle carriers bud from restricted domains of the ER membrane known as transitional ER (tER) or ER Exit Sites (ERESs). At these sites the accessory protein Sec16 initiates the multistep assembly of the COPII coat that is made of five distinct proteins Sar1/Sec23/Sec24/Sec13/Sec31, previously identified in yeast cells by the 2013 Nobel Prize winner Randy W. Schekman (Barlowe et al., 1994; Zanetti et al., 2012). COPII assembly drives vesicle formation, sorting and packaging of cargo molecules and is modulated by different factors according to secretory requirements.
Earlier work by Schekman provided a great contribution for the understanding of the molecular basis of inherited diseases of protein transport and current findings assign an undeniable role to the deficiency of COPII function in human diseases. Indeed, mutations in genes encoding Sec23A COPII-protein lead to un-secreted pro-collagen accumulating within the ER and, consequently, to collagen deposition defects and to skeletal and developmental alterations, as revealed in a rare genetic disease known as skull-Lenticular-Sutural Dysplasia (LSD) (Boyadjiev et al., 2006). Similarly, pre-chylomicrons, large lipid particles, accumulate into the ER of enterocytes when the COPII component SAR1B is mutated, thus causing the Chilomicron Retention Disease (CRD) (Jones et al., 2003).
Therefore, defective COPII function affects large size cargo secretion, suggesting that COPII must have the ability to adapt vesicle coating to the different size or shape of cargo molecules.
As a matter of fact, standard COPII coats are rigid cuboctahedral cages, 60–80 nm in diameter. Thus, COPII vesicles are too little to accommodate large cargo molecules such as pro-collagen, which assembles into elongated bars 300-400 nm long, or chilomicrons ranging from 150 to 500 nm in diameter.
A variety of evidences showed that, to obtain different shapes, COPII employs specific coat subunits or particular cargo adaptors. For an example, in yeast, GPI-anchored proteins (GPI-APs) and the multimeric Pma1 complex require the Sec24 COPII-protein paralog, Lst1/Sfb2, to produce larger COPII vesicle (Peng et al., 2000). Collagen transport requires the adaptor protein, TANGO-1 and its partner cTAGE5, which interact with collagen and Sec23/24, thereby helping collagen enrolment into nascent COPII vesicles (Saito et al., 2009).
The mechanism that makes COPII able to enlarge its size remains still mysterious. However, recent findings show that the ubiquitin ligase Cul3 and its adaptor protein Klhl12 are necessary for COPII-dependent export of pro-collagen suggesting an interesting model to explain this key event in protein trafficking (Jin et al., 2012). Cul3/Klhl12 mono-ubiquitinates Sec31 by targeting alternative lysine residues of the COPII-protein. Although it is not known how ubiquitination affects COPII-size or structure, it is certain that the activity of Cul3/Klhl1 is sufficient to obtain enlargement of COPII as is shown by the overexpression of Klhl12, which generates huge COPII structures and greatly enhance pro-collagen exit from the ER. Certainly, Cul3/Klhl1 is not necessary for the export of smaller or more flexible molecules, such as fibronectin, EGF receptor or β1-integrin suggesting a pro-collagen specific activity for Cul3/Klhl1.
The Cul3/Klhl1 model gives rise to very interesting questions. First of all, Sec31 might not be the only substrate of Cul3/Klhl1, which might influence COPII size by targeting a different COPII subunits. In addition, it is not known whether or not Cul3/Klhl1 could modulate COPII polymerization to help packaging of other large cargo molecules such as lipoproteins and chylomicrons.
All in all, the discovery of Sec31 mono-ubiquitination suggests a very exciting model for the regulation of COPII flexibility and function and opens up new perspectives in the field of vesicular transport.
Bibliography
- Barlowe, C., L. Orci, T. Yeung, M. Hosobuchi, S. Hamamoto, N. Salama, M.F. Rexach, M. Ravazzola, M. Amherdt, and R. Schekman. 1994. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 77:895-907.
- Boyadjiev, S.A., J.C. Fromme, J. Ben, S.S. Chong, C. Nauta, D.J. Hur, G. Zhang, S. Hamamoto, R. Schekman, M. Ravazzola, L. Orci, and W. Eyaid. 2006. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat Genet. 38:1192-1197.
- Gillon, A.D., C.F. Latham, and E.A. Miller. 2012. Vesicle-mediated ER export of proteins and lipids. Biochim Biophys Acta. 1821:1040-1049.
- Jin, L., K.B. Pahuja, K.E. Wickliffe, A. Gorur, C. Baumgartel, R. Schekman, and M. Rape. 2012. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 482:495-500.
- Jones, B., E.L. Jones, S.A. Bonney, H.N. Patel, A.R. Mensenkamp, S. Eichenbaum-Voline, M. Rudling, U. Myrdal, G. Annesi, S. Naik, N. Meadows, A. Quattrone, S.A. Islam, R.P. Naoumova, B. Angelin, R. Infante, E. Levy, C.C. Roy, P.S. Freemont, J. Scott, and C.C. Shoulders. 2003. Mutations in a Sar1 GTPase of COPII vesicles are associated with lipid absorption disorders. Nat Genet. 34:29-31.
- Peng, R., A. De Antoni, and D. Gallwitz. 2000. Evidence for overlapping and distinct functions in protein transport of coat protein Sec24p family members. J Biol Chem. 275:11521-11528.
- Saito, K., M. Chen, F. Bard, S. Chen, H. Zhou, D. Woodley, R. Polischuk, R. Schekman, and V. Malhotra. 2009. TANGO1 Facilitates Cargo Loading at Endoplasmic Reticulum Exit Sites. Cell. 136:891-902.
- Zanetti, G., K.B. Pahuja, S. Studer, S. Shim, and R. Schekman. 2012. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 14:20-28.