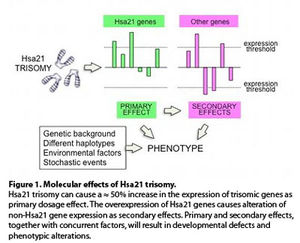
Which are the molecular mechanisms that cause the Down syndrome (DS) phenotype? The most popular hypothesis is that the extra copy of each Hsa21 gene results in an increase in transcription (primary dosage effect) and, as a consequence of that, several genes located on different chromosomes are dysregulated (secondary effect). Primary and secondary effects, together with other concurring factors (Figure 1), determine the final phenotype.
The advent of transcriptome analyses has made it possible to test this model. A comprehensive meta-analysis from 45 studies (1) confirmed the dosage effect hypothesis in DS, and identified 77 Hsa21 genes and 247 non-Hsa21 genes with consistent primary and secondary dysregulation across the studies.
With respect to the first question, a variety of mitochondrial alterations were described in the literature in cells and tissues from DS subjects and from DS mouse models. More recent studies assessed that the overall mitochondrial energy production apparatus is less efficient in DS fetal fibroblasts (4, 5).
This feature is more pronounced in trisomy 21 fibroblasts derived from fetuses with cardiopathy (6) suggesting that mitochondrial dysfunction might concur to generate a more severe cardiac phenotype. Can this concept be extended to other phenotypic traits?
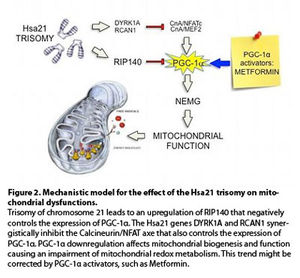
If this overall hypothesis (Figure 2) holds true, a way to correct the mitochondrial dysfunction in DS patients is at hands, since already tested drugs are available that regulate the function of PGC-1α or of its downstream effectors. Among these there is metformin, a widely used anti-diabetic drug. What makes this proposal exiting is the recent finding that metformin promotes neurogenesis in mice and enhances spatial memory formation (8). Could it affect the DS neurological phenotype?
More than 200 protein-coding genes are present on Hsa21 and we have considered here only one of them. Therefore there is still a long way to go. However, even the longest trip starts with one small step.
References
- Vilardell M, Rasche A, Thormann A, Maschke-Dutz E, Pérez-Jurado LA, Lehrach H, Herwig R. 2011. Meta-analysis of heterogeneous Down Syndrome data reveals consistent genome-wide dosage effects related to neurological processes. BMC Genomics 12:229.
- Mao R, Wang X, Spitznagel EL Jr, Frelin LP, Ting JC, Ding H, Kim JW, Ruczinski I, Downey TJ, Pevsner J. 2005. Primary and secondary transcriptional effects in the developing human Down syndrome brain and heart. Genome Biol 6:R107.
- Conti A, Fabbrini F, D'Agostino P, Negri R, Greco D, Genesio R, D'Armiento M, Olla C, Paladini D, Zannini M, Nitsch L. 2007. Altered expression of mitochondrial and extracellular matrix genes in the heart of human fetuses with chromosome 21 trisomy. BMC Genomics 8:268-282.
- Valenti D, Tullo A, Caratozzolo MF, Merafina RS, Scartezzini P, Marra E and Vacca RA. 2010. Impairment of F1F0-ATPase, adenine nucleotide translocator and adenylate kinase causes mitochondrial energy deficit in human skin fibroblasts with chromosome 21 trisomy. Biochem J 431:299-310.
- Valenti D, Manente GA, Moro L, Marra E and Vacca RA. 2011. Deficit of complex I activity in human skin fibroblasts with chromosome 21 trisomy and overproduction of reactive oxygen species by mitochondria: involvement of the cAMP/PKA signalling pathway. Biochem J 435:679-688.
- Piccoli C, Izzo A, Scrima R, Bonfiglio F, Manco R, Negri R, Quarato G, Cela O, Ripoli M, Prisco M, Gentile F, Calì G, Pinton P, Conti A, Nitsch L, Capitanio N. 2013. Chronic pro-oxidative state and mitochondrial dysfunctions are more pronounced in fibroblasts from Down syndrome foeti with congenital heart defects. Hum Mol Genet 22:1218-32.
- Gardiner K. 2006. Transcriptional dysregulation in Down syndrome: predictions for altered protein complex stoichiometries and post-translational modifications, and consequences for learning/behavior genes ELK, CREB, and the estrogen and glucocorticoid receptors. Behav Genet 36:439-453.
- Wang J, Gallagher D, DeVito LM, Cancino GI, Tsui D, He L, Keller GM, Frankland PW, Kaplan DR, Miller FD. 2012. Metformin activates an atypical PKC-CBP pathway to promote neurogenesis and enhance spatial memory formation. Cell Stem Cell 11:23-35.